References
- J. W. Ponder, D. A. Case, "Force Fields for Protein Simulations", Advances in Protein Chemistry, 66, 27-85.
- A. D. MacKerell et al., "All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins", The Journal of Physical Chemistry B, 102, 3586-3616.
- L. D. Schuler, X. Daura, W. F. van Gunsteren, "An improved GROMOS96 force field for aliphatic hydrocarbons in the condensed phase", J. Comput. Chem., 22, 1205-1218.
- W. L. Jorgensen, D. S. Maxwell, J. Tirado-Rives, "Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids". J. Am. Chem. Soc. 118, 11225–11236.
- S. Riniker, "Fixed-Charge Atomistic Force Fields for Molecular Dynamics Simulations in the Condensed Phase: An Overview", Journal of Chemical Information and Modeling, 58 , 565-578.
- T. A. Halgren, W. Damm, "Polarizable force fields", Current Opinion in Structural Biology, 11, 236-242.
- N. Gresh et al., "Anisotropic, Polarizable Molecular Mechanics Studies of Inter- and Intramolecular Interactions and Ligand-Macromolecule Complexes. A Bottom-Up Strategy", Journal of Chemical Theory and Computation, 3, 1960–1986.
- "Anthony Stone: Computer programs". www-stone.ch.cam.ac.uk
- J. R. Maple et al., "A Polarizable Force Field and Continuum Solvation Methodology for Modeling of Protein-Ligand Interactions", Journal of Chemical Theory and Computation, 1, 694–715.
- J. Gao, D. Habibollazadeh, L. Shao, "A polarizable intermolecular potential function for simulation of liquid alcohols". The Journal of Physical Chemistry, 99, 16460–7.
- L. Yang et al., "New-generation amber united-atom force field", The Journal of Physical Chemistry B, 110, 13166–76.
- S. Patel, C. L. Brooks, "CHARMM fluctuating charge force field for proteins: I parameterization and application to bulk organic liquid simulations", Journal of Computational Chemistry, 25, 1–15.
- J. Barnoud, L. Monticelli, (2015) "Coarse-Grained Force Fields for Molecular Simulations. In: Kukol A. (eds) Molecular Modeling of Proteins. Methods in Molecular Biology (Methods and Protocols)", vol 1215. Humana Press, New York, NY.
- J. Barnoud, L. Monticelli, (2015) "Coarse-Grained Force Fields for Molecular Simulations. In: Kukol A. (eds) Molecular Modeling of Proteins. Methods in Molecular Biology (Methods and Protocols)", vol 1215. Humana Press, New York, NY.
- S. J. Marrink et al., "The MARTINI force field: coarse grained model for biomolecular simulations". The Journal of Physical Chemistry B., 111, 7812–24.
- D. Leonardo et al.,"SIRAH: A Structurally Unbiased Coarse-Grained Force Field for Proteins with Aqueous Solvation and Long-Range Electrostatics", Journal of Chemical Theory and Computation, 11, 723-739.
- A. Korkut, W. A. Hendrickson, "A force field for virtual atom molecular mechanics of proteins", Proceedings of the National Academy of Sciences of the United States of America, 106, 15667–72.
- Español, P. Warren, "Statistical Mechanics of Dissipative Particle Dynamics", Europhysics Letters, 30, 191–196.
- Senftle et al. "The ReaxFF reactive force-field: development, applications and future directions". npj Comput Mater 2, 15011.
- Warshel, R. M. Weiss, "An empirical valence bond approach for comparing reactions in solutions and in enzymes", Journal of the American Chemical Society, 102, 6218-6226.
- C. T. van Duin, S. Dasgupta, F. Lorant, W. A. Goddard, "ReaxFF: A Reactive Force Field for Hydrocarbons", The Journal of Physical Chemistry A, 105, 9396-9409.
- Behler, M, Parrinello, "Generalized Neural-Network Representation of High-Dimensional Potential-Energy Surfaces", Phys. Rev. Lett. 98, 146401.
- V. Botu et al., "Machine Learning Force Fields: Construction, Validation, and Outlook, The Journal of Physical Chemistry C", 121, 511-522.
- J. Behler et al., "Perspective: Machine learning potentials for atomistic simulations", J. Chem. Phys., 145, 170901.
- D. Rapaport, "The Art of Molecular Dynamics Simulation" (2nd ed.), Cambridge: Cambridge University Press.
- H. C. Andersen, "Molecular dynamics simulations at constant pressure and/or temperature", J. Chem. Phys. 72, 2384.
- W. G. Hoover, "Canonical dynamics: Equilibrium phase-space distributions", Phys. Rev. A, 31, 1695.
- H. J. C. Berendsen et al., "Molecular dynamics with coupling to an external bath", J. Chem. Phys. 81, 3684-90.
- G. Bussia, D. Donadio, M. Parrinello, "Canonical sampling through velocity rescaling", J. Chem. Phys. 126, 014101.
- H. J. C. Berendsen et al., "Molecular dynamics with coupling to an external bath", J. Chem. Phys. 81, 3684-90.
- M. Parrinello, A. Rahman, "Polymorphic transitions in single crystals: A new molecular dynamics method, Journal of Applied Physics", 52, 7182.
- J. Chen, 2018 IOP Conf. Ser.: Earth Environ. Sci., 128, 012110.
- John von Neumann Inst. for Computing, NIC series 1., IV, 562 S. (2000).

Hi
Hello. And Bye.
Congratulations. Great content.
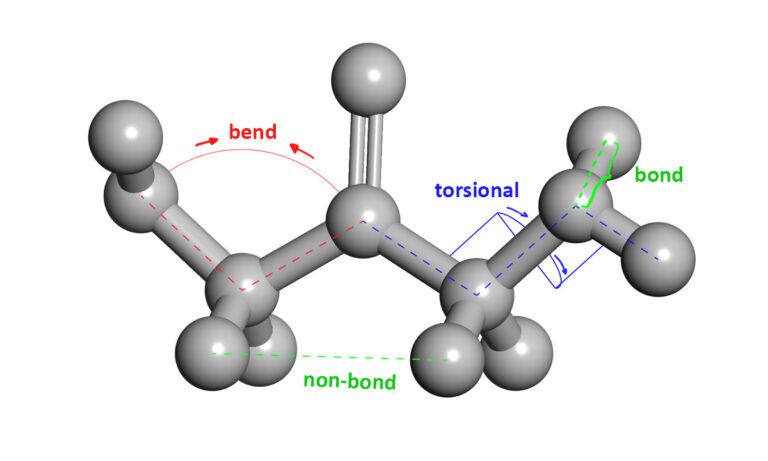
An attractive alternative is to work with an atomic-level computer simulation of the relevant biomolecules. Molecular dynamics MD simulations predict how every atom in a protein or other molecular system will move over time, based on a general model of the physics governing interatomic interactions Karplus and Mc Cammon, 2002. These simulations can capture a wide variety of important biomolecular processes, including conformational change, ligand binding, and protein folding, revealing the positions of all the atoms at femtosecond temporal resolution.
In general, a small thermostat timescale parameter causes a tighter coupling to the virtual heat bath and a system temperature that closely follows the reservoir temperature. Such a tight thermostat coupling is typically accompanied by a more pronounced interference with the natural dynamics of the particles, however. Therefore, when aiming at a precise measurement of dynamical properties, such as diffusion or vibrations, you should use a larger value of the thermal coupling constant or consider running the simulation in the NVE ensemble to avoid artifacts of the thermostat.
Bravo!!
I just want to say thank you for this great website. I found a solution here on insilicosci.com for my issue.