We Can Help You with Your Studies through in-silico methods
If you’re looking to outsource in silico projects, consider INSILICOSCI services. We utilise computer simulations and modelling to save you both resources and time. Our team has years of expertise in computational techniques and is committed to producing high-quality outcomes that meet your requirements. We welcome in-silico projects of various scales and complexity, and we are delighted to assist you with your in silico projects. The four primary areas of our expertise are:
Computational Chemistry methods for molecular compounds
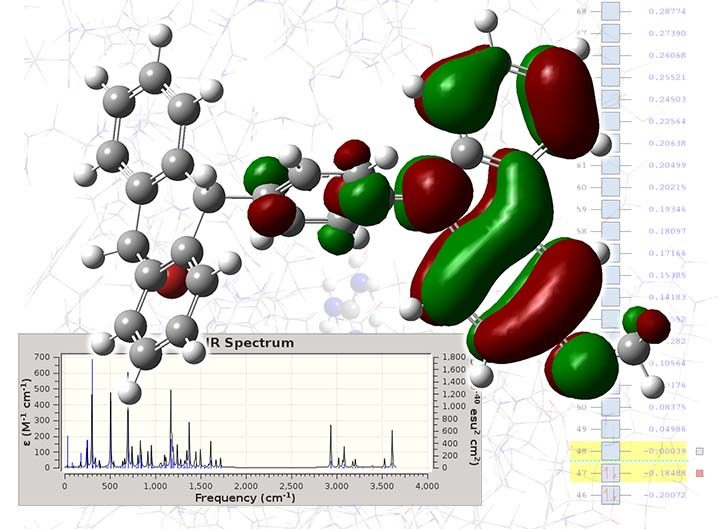
Computational chemistry encompasses a wide range of in-silico techniques, from semi-empirical and DFT to high-level post-SCF methods. In computational chemistry, we mostly deal with molecular compounds, and the key problem is to solve the Schrodinger equation for the electronic system. By solving the equation, we get a clear description of the molecule’s structure, and by analysing the wave function, or electron density, we determine various chemical properties. Computational chemistry also can predict chemical reactions and investigate their mechanism kinetics.
Here in INSILICOSCI, we use a variety of computational chemistry software, including Gaussian, NWChem, Orca, Materials Studio, and GAMESS US.
Density functional theory for solid-state materials
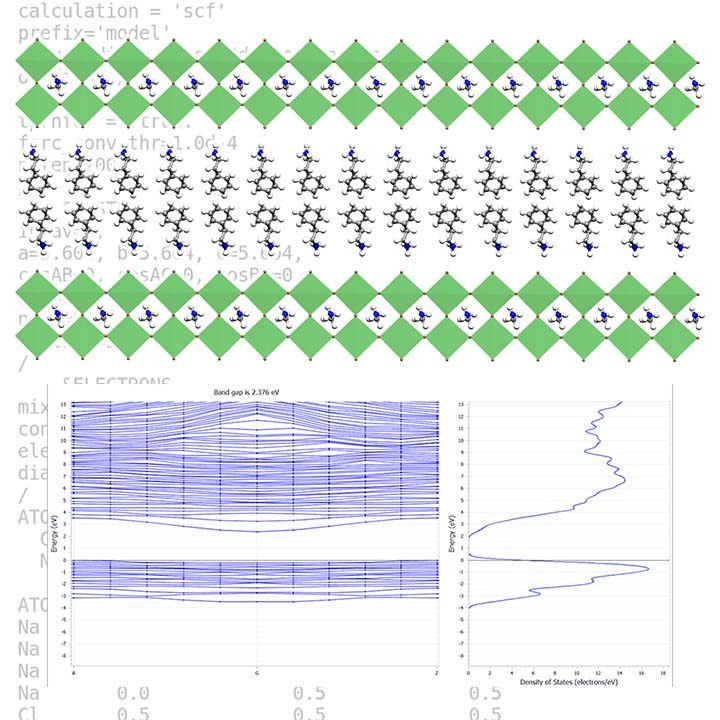
Nowadays, we use density functional theory in many science fields, but the most successful application of DFT is to investigate solid-state materials. Current DFT codes can calculate a wide range of solid-state material structural, chemical, optical, spectroscopic, elastic, vibrational, and thermodynamic properties. DFT calculations can now predict a considerable portion of the properties for a material, not only under ambient conditions but also in extremely high-pressure conditions. It is also possible to study solid-state phenomena by simulating the evolution of the material structure over a few hundred picoseconds. This feature lets us explore the phenomena’ dynamics and kinetics and calculate their thermodynamic properties.
To tackle solid-state problems, our group utilises a set of plane-wave DFT codes, including VASP, quantum-espresso, ABINIT, and CASTEP.
Monte Carlo and molecular dynamics simulations
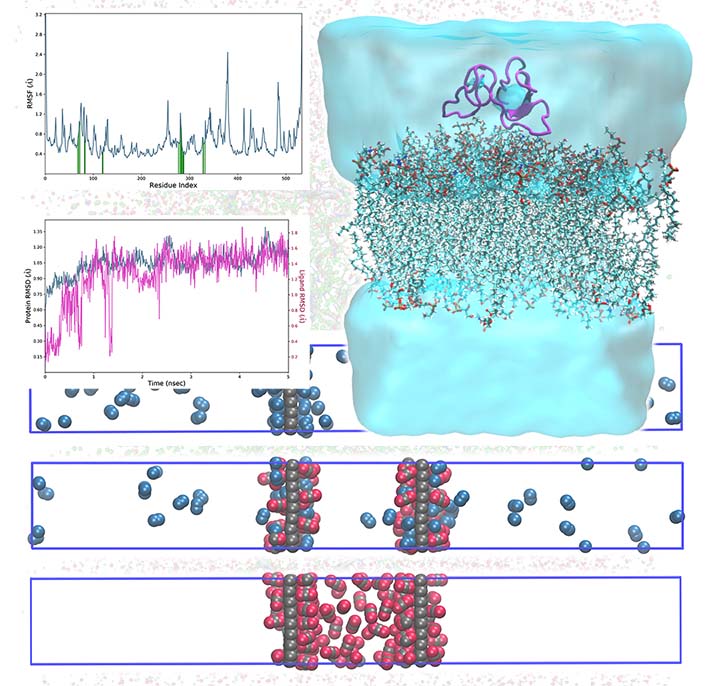
Monte Carlo and Molecular Dynamics simulations are among the most effective computational tools and have numerous applications in physics, chemistry, biochemistry, and materials science. In these simulation techniques, we set up an initial array of atoms and then let the simulation generate an ensemble of representative configurations under specific thermodynamic conditions.
We can explore many biological and non-biological phenomena using MC/MD simulations, including protein-ligand interaction, drug-carrier interaction, mutation, drug delivery across the cell membrane, micellization, self-assembly, deformation, dislocation, separation, and adsorption.
Our arsenal of simulation tools includes GROMACS, Materials Studio, LAMMPS, NAMD, and Schrodinger software packages.
in silico drug design and drug discovery
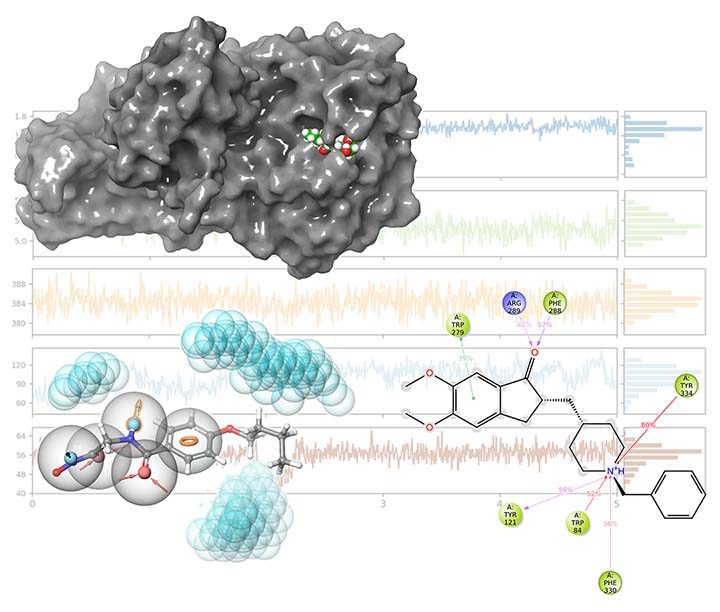
Today, researchers have the luxury of employing in-silico drug discovery techniques to reduce or completely avoid the usage of exhausting, costly and time-consuming traditional drug discovery approaches. We can easily filter thousands of candidate compounds and find potential drugs among them using techniques like pharmacophore modelling, virtual screening, and molecular docking. Using regression techniques like 3D-QSAR, we can also estimate chemical compounds’ biological activity against a target. Finally, molecular dynamics techniques provide us with a vivid insight into the mechanism of the action of drugs.
Insilicosci team uses Schrodinger, Materials Studio, GROMACS and AutoDock packages to help you with your drug discovery and design projects.
ab initio molecular dynamics
Classical molecular dynamics simulation is a powerful tool for investigating many biological and non-biological systems, but it has a severe flaw. Classical MD needs pre-defined potentials (force fields) to work, which means we can only perform simulation on a restricted range of systems whose force fields have been parametrised before. To add insult to injury, most force fields cannot handle chemically complex situations, such as systems with many different atom types or systems that experience changes in the covalent banding pattern throughout the simulation. Computational scientists developed Ab initio molecular dynamics (AIMD) techniques to address this issue. The basic idea behind AIMD methods is to calculate the forces acting on nuclei from electronic structure calculations performed “on the fly” during the simulation. This approach eliminates the need for pre-defined force fields, allowing us to perform ab initio molecular dynamics simulations on virtually any molecular system, regardless of how chemically complex it is — however, since AIMD simulations are heavily computationally consuming, we are limited to short time and size scales.
Here in Insilicosci, we can use a software toolset including Gaussian, CPMD, quantum-espresso, VASP, and NWChem to utilize multiple AIMD methods such as Car-Parrinello, Born-Oppenheimer, and atom-centred density matrix propagation molecular dynamics.
types of our services
conduct in-silico projects
You can completely outsource an in-silico project to us through this service, and we will take care of everything from researching to creating molecular structures and input files, running calculations, analysing output files, and extracting results from them. We accept projects in chemistry, biochemistry, material science, drug design, and drug discovery.
computational assist
Sometimes you may encounter a problem with a specific computational task in your in-silico project or you may be an experimental scientist who is interested in incorporating an in silico analysis into your research. Here you can outsource that specific analysis to INSILICOSCI as a computational assist service.
making force-fields and molecular structures
Making a correct force field for the system is one of the most challenging tasks in atomistic simulations. Also, generating molecular structures for some systems is tricky. If you are experiencing difficulties in these areas, you can outsource these tasks to us.
run computational jobs
Via this service, you set up the input files, and we just run your files on our high-performance computing server. This service is suitable for researchers who are proficient in computational methods yet require a powerful computational server. At the moment, we accept input files for GROMACS, NAMD, LAMMPS, Amber, VASP, Quantum Espresso, Abinit, and NWChem software.
How can you request for a service?
Send us an email or a WhatsApp message. Provide as much information about your project or calculation as possible. It is best to send us an article similar to your study and identify which calculations and analysis you need from the article.
We also need to know about the molecular system you’re studying. You can send us molecular structures in any 2D or 3D format, or you can provide a link to a website containing the structure (e.g. PubChem or RCSB). If you’re looking to study any chemical/biochemical reaction or process, please specify them clearly. You can simply draw the reaction on the paper and take a photo of it or refer to an article or website that describes the reaction.
If you only want us to run your computational jobs on our server, please provide all the input files and test the inputs before sending them to us.
Pricing
Because of the extensive variety of in silico analysis and since molecular systems have very different labour demands. We cannot supply a pricing list. The exact price of your job will be determined after considering the details of your project, and it will be affected by many factors, including preparation and analysis time and computing cost.
We will not seek any interest in your studies, but in some cases you can get a discount by mentioning our contributions to the publication.
